PathoAI Cockpit - Pathology Data Science for Molecular and Digital Pathology AI
Results of this project are explained in the final report.
- Sponsored by: TUM AI for Pathology
- Project Lead: Dr. Ricardo Acevedo Cabra
- Scientific Lead: Prof. Dr. Peter Schüffler, PD Dr. Katja Steiger and Dipl.-Biol. Nicole Pfarr
- TUM Co-Mentor: TBA
- Term: Summer semester 2023
- Application deadline 29.01.2023
Apply to this project here
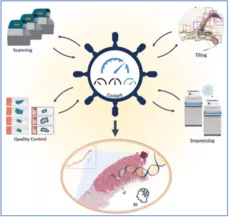
tasks in digitization and for compiling relevant datasets used in pathology AI.
Artificial intelligence (AI) is a powerful tool to detect diseases and genetic alterations in digital histology1. E.g., state-of-the-art deep-learning (DL) models have not only proven to discover small lesions of cancer in digital slides2, but have also shown strong capabilities to detect genetic variations3 and mutations4 of various genes in different kinds of cancer, drastically enhancing the specificity for molecular tests. However, the development of such AI models requires large data sets of digital tissue samples associated with their molecular data.
The TUM established molecular and digital pathology workflows in 2019 and 2021, respectively. However, data are currently unstructured, poorly related to each other and not necessary of the required quality, impeding the fast and simple compilation of relevant data sets for AI in pathology.
Therefore, we aim to build a centralized Cockpit for Pathology AI. This cockpit will show relevant statistics of our current data set, correlating molecular with digital pathology data, and provides a comprehensive toolbox for real-time data processing and quality control. Ultimately, the cockpit will help AI researchers to search for relevant and corelated data sets with required quality. In this call, we concentrate on the digital pathology part and the quality control of digital slides.
Goals
- Develop a dashboard for digital pathology, indicating current digitization rates, errors and problems during scanning, overview of diseases and entities in the digital database. For this, write interfaces to connect to different pathology databases such as Nexus LIS and eSlideManager. Bonus: Extending the dashboard with information from molecular pathology: Current sequencing rate of cases, number of sequenced cases and correlated digitized cases. Requirements: Software project 1 will develop a frontend/backend server application. Interest in coding (frontend, backend) and database management is expected.
- Develop an algorithm for the automatic quantification of post-scan quality of slides, regarding unscanned tissue and touching tissue, using AI and/or image processing
- Compare existing (AI) algorithms for the automatic quantification of post-scan quality of slides, regarding blurriness, pen and airbubbles. AI exists in literature. Optionally extend/improve AI.
- Develop an algorithm for the automatic quantification of post-scan quality of slides, regarding striping errors using AI and/or image processing. Requirements: AI projects 2, 3, 4 can be developed from scratch using python and ML or using existing toolboxes such as PathML, TIA Toolbox or others. Interest in ML, scripting and working on the LRZ Compute Cluster are expected.
- Students from all projects are expected to elaborate together an appropriate data format for AI output which is ingestible in the dashboard (e.g., json, xml, file-based or other).
We currently digitize 200.000 slides per year. A digital slide is a high-resolution image of processed tissue exceeding 100k x 100k px, thus requiring special handling and processing. The cockpit affects all data. However, for development of induvial AI models, subsets of data will be selected which make sense for the particular task.
References
- Echle A, Rindtorff NT, Brinker TJ, Luedde T, Pearson AT, Kather JN. Deep learning in cancer pathology: a new generation of clinical biomarkers. Br J Cancer. Published online November 18, 2020:1-11. doi:10.1038/s41416-020-01122-x
- Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25(8):1301-1309. doi:10.1038/s41591-019-0508-1
- Yamashita R, Long J, Longacre T, et al. Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol. 2021;22(1):132-141. doi:10.1016/S1470-2045(20)30535-0
- Bilal M, Raza SEA, Azam A, et al. Development and validation of a weakly supervised deep learning framework to predict the status of molecular pathways and key mutations in colorectal cancer from routine histology images: a retrospective study. Lancet Digit Health. 2021;3(12):e763-e772. doi:10.1016/S2589-7500(21)00180-1
Important notice
Accepted students to this project should attend online workshops at the LRZ in April 2023 before the semester starts, unless they have proven knowledge. More information will be provided to students accepted to this project.