Topology regularised foundation model for medical image segmentation
Results of this project are shown in the demo video and the final report (PDF) below:
- Sponsored by: TUM Chair of Mathematical Modeling of Biological Systems (MDSI Prof. Theis) and Helmholtz AI
- Project Lead: Dr. Ricardo Acevedo Cabra
- Scientific Lead: Dr. Tingying Peng, Dr. Bastian Rieck, Dr. Carsten Marr
- TUM Co-Mentor: Dr. Alessandro Scagliotti
- Term: Winter semester 2022
- Application deadline: Sunday 23.07.2023
Background & Motivation
In the broader AI fields of natural language processing and computer vision, transformer-based large language models have demonstrated promising results across a wide range of mono-modal and multi-modal applications, ultimately aiming to achieve what is known as artificial general intelligence (AGI). Notable examples of these endeavours include ChatGPT, Facebook’s Segment Anything Model (SAM1), and Contrastive Language-Image Pre-Training (CLIP)2. These models, trained on vast amounts of data, generate robust and adaptable representations, enabling them to serve as a foundation for various downstream tasks and applications (hence the term “Foundation model”). In this project, we explore the potential of applying these models, particularly SAM, in the medical image domain for segmentation.
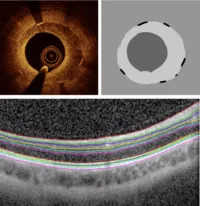
In ongoing research projects Helmholtz Munich, we have collected intravascular optical coherence tomography (OCT) data in collaboration with the German Heart Center Munich and retinal OCT data from the LMU eye clinics. Conventionally, a nnUnet is trained to segment regions of interest, e.g. different tissue types in intravascular OCT, (see Figure 1 top) or different retinal layers (shown in Figure 1 bottom). However, these models struggle when handling hitherto-unknown image types, or data from different imaging centres. We would like to leverage SAM in this context and evaluate its segmentation performance on medical images.
Objectives:
- Plugin Development and Model Integration: the first objective is to create a plugin that allows for the application of the SAM in the medical imaging domain. This involves understanding SAM's design and mechanisms and adapting it for medical imaging use, particularly with intravascular and retinal OCT data.
- Performance Evaluation and Prompt Optimization: the next objective is to evaluate and optimise SAM's effectiveness in the medical imaging context. This includes a) gauging SAM's segmentation performance on medical images, especially in comparison to existing methods like nnUnet, and b) investigating the impact of various types of prompts (text, points, boxes) on the model's output quality.
- Advanced Model Enhancement and Knowledge Transfer: The most complex objective involves implementing advanced techniques to further enhance the model's capabilities and ensure effective knowledge transfer. This incorporates a) integrating novel loss terms based on recent advances in geometrical and topological representation learning to enhance the model's predictive and generalization performance, b) facilitating multi-scale learning c) testing the model's adaptability to unseen data from different imaging centres.
Requirements:
Solid mathematic background, prior knowledge of machine learning / deep learning and Python coding skills with experience in Pytorch or Tensorflow.