Spatial and Functional Dynamics of Oligodendrocytes in Alzheimer's Disease
- Sponsored by: TUM Mathematical Modelling of Biological Systems & Helmholtz Munich
- Project lead: Dr. Ricardo Acevedo Cabra
- Scientific lead: PhD Candidates Lisa Evangelista and Francesca Drummer
- TUM co-mentor: TBA
- Term: Winter semester 2025
- Application deadline: Sunday 20.07.2025
Apply to this project here
Background
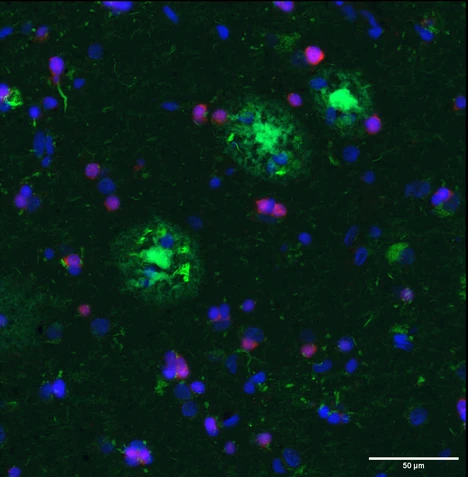
Alzheimer's disease (AD) is a progressive neurodegenerative disease of the central nervous system (CNS) characterized by the accumulation of extracellular amyloid-beta peptide (Aβ) plaques and neurofibrillary tangles, which cause neuroinflammation, synaptic loss and eventually neuronal loss, ultimately resulting in dementia. [1]. For decades, AD has been studied mainly from a neuronal point of view. In recent years, however, the latest advances in the field have clearly demonstrated glial involvement in disease pathogenesis and progression. Oligodendrocytes, the myelin forming cells in the CNS, are the first cell type to transcriptionally change in the earliest stages of the disease [2], moreover, they have been shown to significantly contribute to the production of Aß in the brain [3]. Importantly, oligodendrocytes are not a homogeneous cell population, in fact distinct states of mature oligodendrocytes exist and they are considered to have different functional roles [4] [5]. The selective loss or the dysfunction of some of these mature states in AD could explain the regional vulnerability characteristic of the disease [unpublished data]. Although great progress has been made towards defining cell states in the CNS using single-cell transcriptomics, these approaches do not preserve spatial information regarding the distribution of oligodendroglial cells in the different cortical layers and in the white matter nor their relationship with Aß plaques. The aim of this project is to explore the response of cortical and white matter oligodendrocytes in the human brain and to unravel how their functional changes contribute to the pathogenesis of Alzheimer’s disease.
Data
We are working with two publicly available human spatial transcriptomics (ST) datasets from the middle temporal gyrus (MTG): 1) imaging based ST technology - multiplexed error-robust fluorescence in situ-hybridization (MERFISH) - from 24 donors [6] and 2) a sequencing based - Visium - dataset from 12 donors [7]. Both datasets have donors from different AD pathology states.
Tasks
- Data exploration and preprocessing: Before using the ST data for downstream evaluation we need to understand the quality of the data and difference between ST technologies (spatial resolution vs gene detection). This includes e.i. evaluating how comparable the technologies are: What common genes are measured between technologies? Do the slides show similar cell distribution? Furthermore, we will extend the data annotations with niche and region labels using existing methods (e.i. CellCharter [8], BANKSY [9]).
- Phenotype differences: Before we zoom in to cellular interactions we evaluate how cellular organization and tissue architecture differ across AD pathologies using existing tools such as GraphCompass [10] & Kontextual [11].
- Cell interactions across conditions: Lastly, we are interested in evaluating interactions across length scales in the tissue environment. We will apply a novel tool (InterScale) developed in the Theis lab to understand local and global interactions across and within the white matter
References
[1] Wilson, David M. et al. Hallmarks of neurodegenerative diseases. Cell, Volume 186, Issue 4, 693 - 714
[2] Cain, A., Taga, M., McCabe, C. et al. Multicellular communities are perturbed in the aging human brain and Alzheimer’s disease. Nat Neurosci 26, 1267–1280 (2023). doi.org/10.1038/s41593-023-01356-x
[3] Sasmita, A.O., Depp, C., Nazarenko, T. et al. Oligodendrocytes produce amyloid-β and contribute to plaque formation alongside neurons in Alzheimer’s disease model mice. Nat Neurosci 27, 1668–1674 (2024). doi.org/10.1038/s41593-024-01730-3
[4] Jäkel, S., Agirre, E., Mendanha Falcão, A. et al. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566, 543–547 (2019). https://doi.org/10.1038/s41586-019-0903-2
[5] Falcão, A.M., van Bruggen, D., Marques, S. et al. Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat Med 24, 1837–1844 (2018). doi.org/10.1038/s41591-018-0236-y
[6] Gabitto, M.I., Travaglini, K.J., Rachleff, V.M. et al. Integrated multimodal cell atlas of Alzheimer’s disease. Nat Neurosci 27, 2366–2383 (2024). doi.org/10.1038/s41593-024-01774-5
[7] Chen S, Chang Y, Li L, Acosta D, Li Y, Guo Q, Wang C, Turkes E, Morrison C, Julian D, Hester ME, Scharre DW, Santiskulvong C, Song SX, Plummer JT, Serrano GE, Beach TG, Duff KE, Ma Q, Fu H. Spatially resolved transcriptomics reveals genes associated with the vulnerability of middle temporal gyrus in Alzheimer's disease. Acta Neuropathol Commun. 2022 Dec 21;10(1):188. doi: 10.1186/s40478-022-01494-6. PMID: 36544231; PMCID: PMC9773466.
[8] Varrone, M., Tavernari, D., Santamaria-Martínez, A., Walsh, L. A. & Ciriello, G. CellCharter reveals spatial cell niches associated with tissue remodeling and cell plasticity. Nat Genet 56, 74–84 (2024).
[9] Singhal, V. et al. BANKSY unifies cell typing and tissue domain segmentation for scalable spatial omics data analysis. Nat Genet 56, 431–441 (2024).
[10] Ali, M. et al. GraphCompass: Spatial metrics for differential analyses of cell organization across conditions. 2024.02.02.578605 Preprint at doi.org/10.1101/2024.02.02.578605 (2024).
[11] Ameen, F., Robertson, N., Lin, D. M., Ghazanfar, S. & Patrick, E. Kontextual: Reframing analysis of spatial omics data reveals consistent cell relationships across images. 2024.09.03.611109 Preprint at doi.org/10.1101/2024.09.03.611109 (2024).
Apply to this project here