Partial 3D Thyroid Registration
This project took place in summer term 2022, you CAN NOT apply to this project anymore!
Results of this project are explained in detail in the final report.
- Sponsored by: TUM Chair for Computer Aided Medical Procedures & Augmented Reality (CAMP), MDSI Prof. Nassir Navab
- Project Lead: Dr. Ricardo Acevedo Cabra
- Scientific Lead: PhD candidate Mahdi Saleh & PhD candidate Lennart Bastian
- TUM Co-Mentor: Dr. Ricardo Acevedo Cabra
- Term: Summer semester 2022
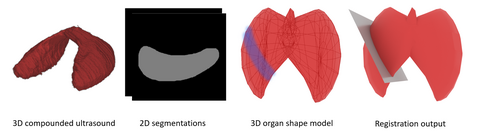
Project Description
Statistical shape models (SSMs) are a well-established tool for mathematically understanding 3D objects. Particularly in the medical context, the last few decades have seen many approaches for modeling human organs with statistical parameterization. SSMs are not only useful for encapsulating normal anatomical variations of an organ, but can also represent pathological cases, anatomical dysmorphisms, or be used to quantify longitudinal variation for individuals (Ambellan et al., 2019). An important decision on statistical parameterization is how to build correspondence between the statistical model and new training cases. Recently Saleh et al. proposed learning geometric landmarks on a mesh, by learning descriptors on the surface with a GCN (Saleh et al., 2020). This project seeks to investigate whether such correspondence learning is possible for a Thyroid dataset captured at Rechts der Isar consisting of 16 paired MRI and 3D Ultrasound scans. Due to its largely rigid structure, and the availability of expert 3D segmentation annotations, the thyroid proves an excellent starting point for deformable shape registration on human organs.
Student tasks
- Literature review. Review several methods commonly used for statistical shape modeling and deformable registration of human organs, particularly the thyroid. To start, see i.e. (Ambellan et al., 2019)
- Explore two thyroid data modalities: MRI and 3D Ultrasound scan. Both modalities have been segmented by radiologists at the Rechts der Isar. The MRI dataset may serve as a better ground truth for statistical model generation.
- Registration with 3D GCNs. Apply the method suggested by (Saleh et al., 2020) for partial registration of individual thyroid segments to their 3D completed shapes. Attempt to register partial matches to a 3D statistical model.
Accepted students to this project should attend (unless they have proven knowledge) online workshops at the LRZ from 19.04. - 22.04.22. More information will be provided to students accepted to this project.
References
Ambellan, F., Lamecker, H., von Tycowicz, C., & Zachow, S. (2019). Statistical Shape Models: Understanding and Mastering Variation in Anatomy. In P. M. Rea (Ed.), Biomedical Visualisation: Volume 3 (pp. 67–84). Springer International Publishing. https://doi.org/10.1007/978-3-030-19385-0_5
Litany, O., Remez, T., Rodolà, E., Bronstein, A. M., & Bronstein, M. M. (2017). Deep Functional Maps: Structured Prediction for Dense Shape Correspondence. ArXiv:1704.08686 [Cs]. http://arxiv.org/abs/1704.08686
Saleh, M., Dehghani, S., Busam, B., Navab, N., & Tombari, F. (2020). Graphite: GRAPH-Induced feaTure Extraction for Point Cloud Registration. ArXiv:2010.09079 [Cs]. http://arxiv.org/abs/2010.09079
